Training the ensemble SVM model to predict cell type directly
Compiled: November 06, 2023
Source:vignettes/misc_disco_pbmc_svm.Rmd
misc_disco_pbmc_svm.RmdProjectSVR also implemented a wrapper
FitEnsemblMultiClassif() to train an ensemble SVM model for
cell type auto-annotation. In this tutorial, we show how to train such a
model and use it for cell type annotation.
Download Related Dataset
library(ProjectSVR)
library(Seurat)
library(tidyverse)
options(timeout = max(3600, getOption("timeout")))
`%notin%` <- Negate(`%in%`)
if (!dir.exists("models")) dir.create("models")
if (!dir.exists("reference")) dir.create("reference")
if (!dir.exists("query")) dir.create("query")
# reference model
download.file(url = "https://zenodo.org/record/8350732/files/model.disco_pbmc.rds",
destfile = "models/model.disco_pbmc.rds")
# reference data
download.file(url = "https://zenodo.org/record/8350746/files/mTCA.seurat.slim.qs",
destfile = "reference/DISCO_hPBMCs.seurat.slim.qs")
# query data
download.file(url = "https://zenodo.org/record/8350748/files/query_hPBMCs.seurat.slim.qs",
destfile = "query/query_hPBMCs.seurat.slim.qs")Build Reference Model
data("pals")
seu.ref <- qs::qread("reference/DISCO_hPBMCs.seurat.slim.qs")
p1 <- DimPlot(seu.ref, pt.size = .4) + scale_color_manual(values = pals$disco_blood)
LabelClusters(p1, id = "ident")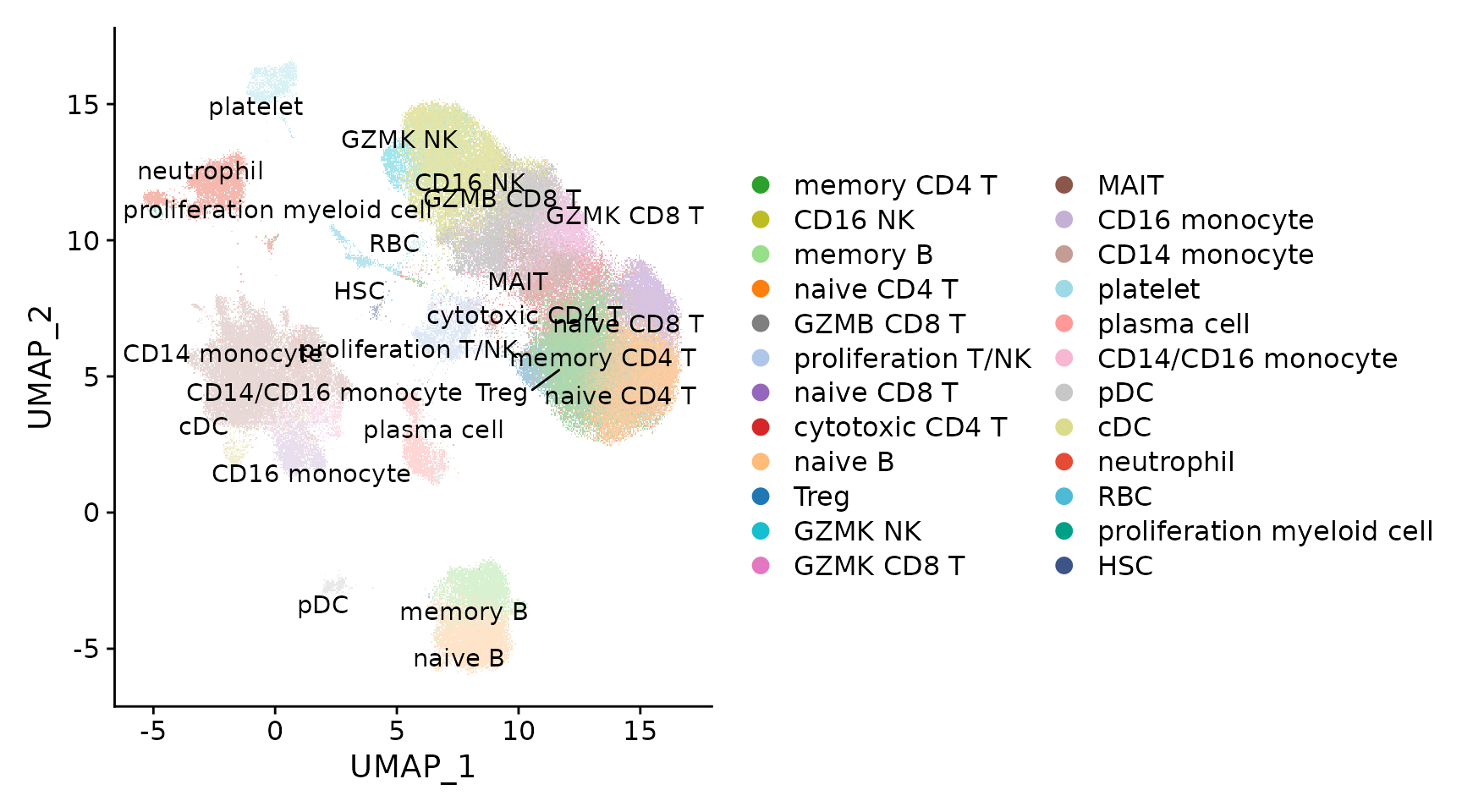
Transfer raw count matrix to gene set score matrix
reference <- readRDS("models/model.disco_pbmc.rds")
top.genes <- reference$genes$gene.sets
bg.genes <- reference$genes$bg.genes
reference$gss.method## [1] "UCell"
seu.ref <- ComputeModuleScore(seu.ref, gene.sets = top.genes, bg.genes = bg.genes, method = "UCell", cores = 5)
# The signature score matrix is stored in 'SignatureScore' assay
Assays(seu.ref)## [1] "RNA" "SignatureScore"
DefaultAssay(seu.ref) <- "SignatureScore"Training reference model
gss.mat <- FetchData(seu.ref, vars = rownames(seu.ref))
cell.types <- FetchData(seu.ref, vars = c("cell_type", "cell_subtype"))
batch.size = 8000 # number of subsampled cells for each SVR model
n.models = 20 # number of SVR models trained
svm.model <- FitEnsemblMultiClassif(feature.mat = gss.mat,
cell.types = cell.types,
batch.size = batch.size,
n.models = n.models,
balance.cell.type = TRUE, # balanced sampling for each cell label
cores = 10)
## save model to reference object
reference$models$cell_type <- svm.model
qs::qsave(reference, "models/model.disco_pbmc.v2.qs")Cell Type Annotation
seu.q <- qs::qread("query/query_hPBMCs.seurat.slim.qs")
## map query
seu.q <- ProjectSVR::MapQuery(seu.q, reference = reference, add.map.qual = T, ncores = 10)
seu.q## An object of class Seurat
## 33718 features across 20886 samples within 2 assays
## Active assay: SignatureScore (24 features, 0 variable features)
## 1 other assay present: RNA
## 3 dimensional reductions calculated: pca.umap, harmony.umap, ref.umap
## predict cell type
gss.mat.q <- FetchData(seu.q, vars = rownames(seu.q))
pred.res <- PredictNewdata(feature.mat = gss.mat.q, model = svm.model, cores = 10)
head(pred.res)## cell_type cell_subtype
## threepfresh_AAACCTGAGCATCATC B cells naive B
## threepfresh_AAACCTGAGCTAACTC monocyte CD14 monocyte
## threepfresh_AAACCTGAGCTAGTGG CD4+ T cells memory CD4 T
## threepfresh_AAACCTGCACATTAGC CD4+ T cells Treg
## threepfresh_AAACCTGCACTGTTAG monocyte CD14 monocyte
## threepfresh_AAACCTGCATAGTAAG cDC cDC
## save results to seurat object
seu.q$cell_type.pred <- pred.res$cell_type
seu.q$cell_subtype.pred <- pred.res$cell_subtype
## visualization
p1 <- DimPlot(seu.q, reduction = "ref.umap", group.by = c("cell_type")) + ggsci::scale_color_d3("category20")
p2 <- DimPlot(seu.q, reduction = "ref.umap", group.by = c("cell_type.pred")) + ggsci::scale_color_d3()
p1 <- LabelClusters(p1, id = "cell_type")
p2 <- LabelClusters(p2, id = "cell_type.pred")
p1 + p2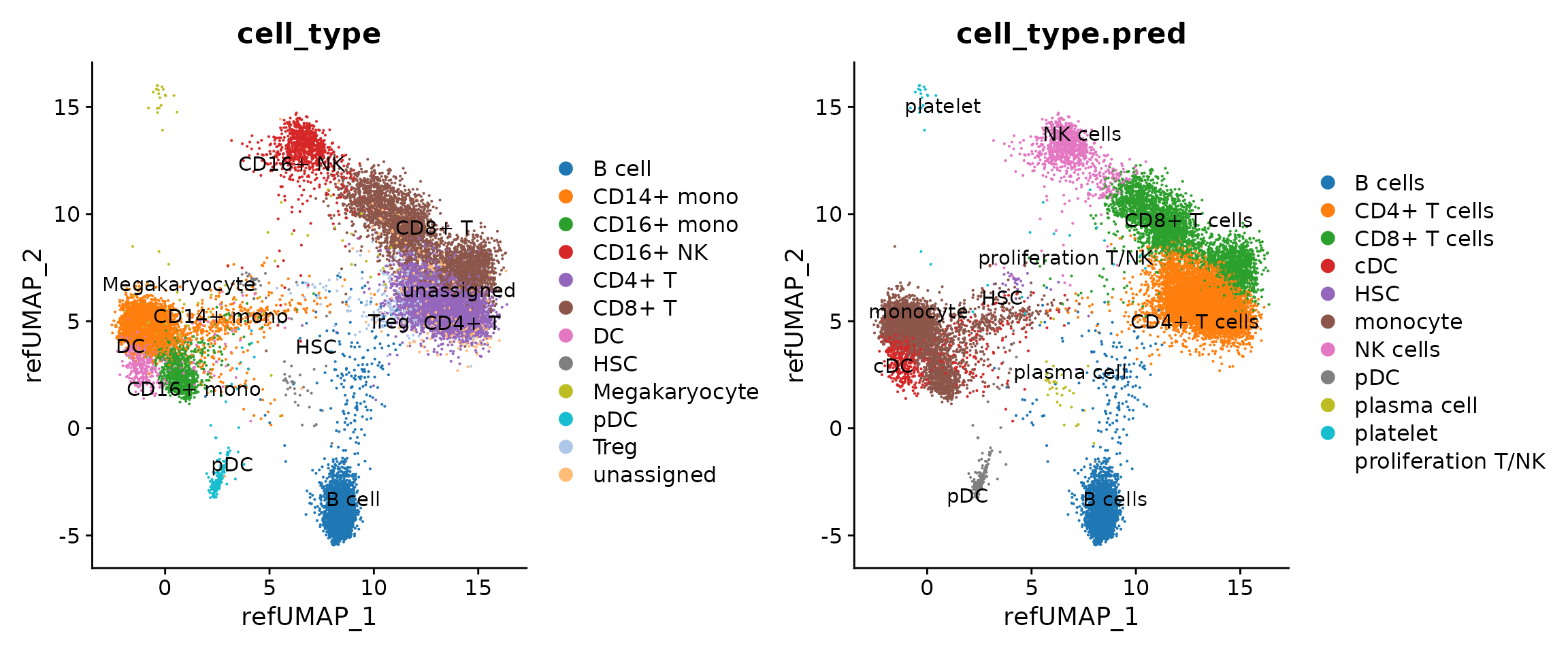
p1 <- DimPlot(seu.q, reduction = "ref.umap", group.by = c("cell_subtype")) + ggsci::scale_color_d3("category20")
p2 <- DimPlot(seu.q, reduction = "ref.umap", group.by = c("cell_subtype.pred")) + scale_color_manual(values = pals$disco_blood)
p1 <- LabelClusters(p1, id = "cell_subtype")
p2 <- LabelClusters(p2, id = "cell_subtype.pred")
p1 + p2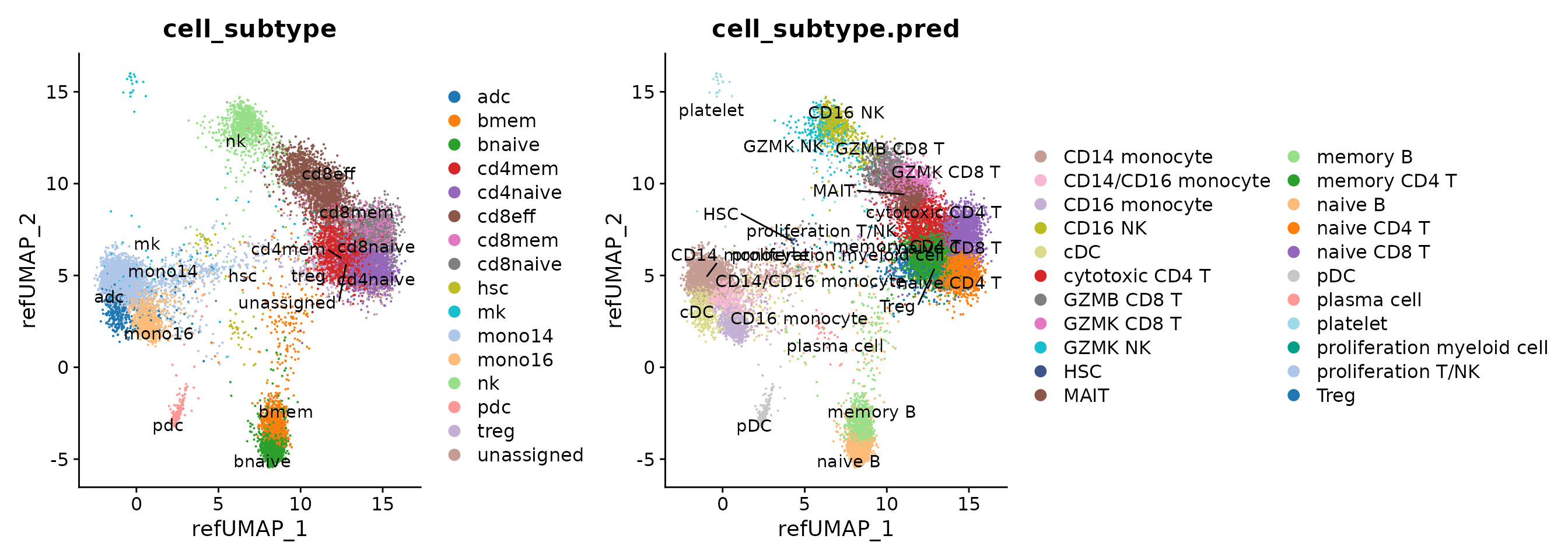
The ProjectSVR gives pretty good cell type predictions. It is quite interesting that the ProjectSVR predicts GZMK+ NK cells. To verify this, we check a marker combination and found that GZMK+ NK cells can be defined by NKG7+/GZMK+/CD16-/CD8A-, which supports the ProjectSVR’s prediction.
DefaultAssay(seu.q) <- "RNA"
seu.q[["RNA"]]@counts <- seu.q[["RNA"]]@data
seu.q <- NormalizeData(seu.q)
FeaturePlot(seu.q, reduction = "ref.umap", features = c("GZMK", "FCGR3A", "NKG7", "CD8A"), ncol = 2)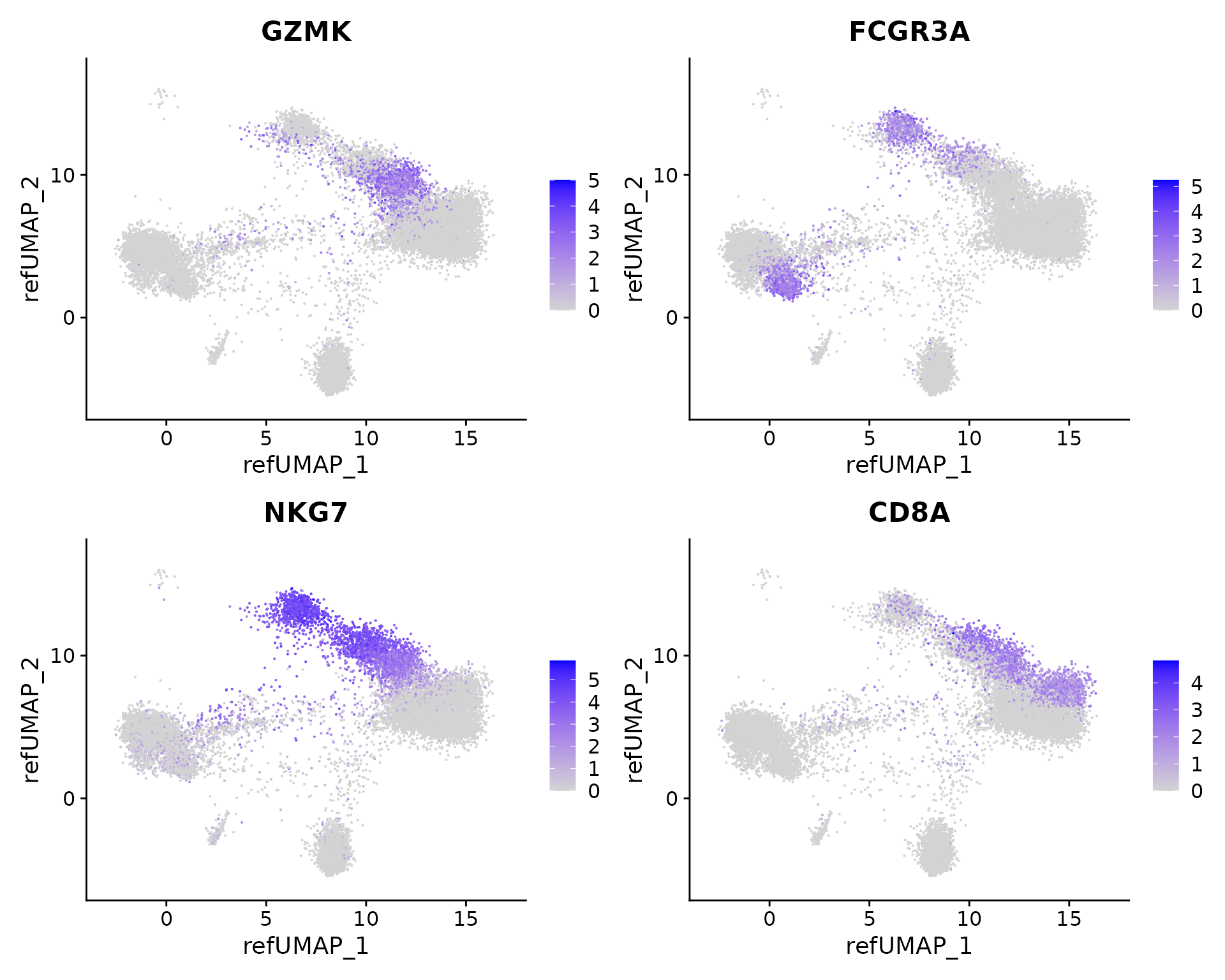
Session Info
## R version 4.1.2 (2021-11-01)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3
## [5] purrr_1.0.2 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
## [9] ggplot2_3.4.3 tidyverse_2.0.0 SeuratObject_4.1.3 Seurat_4.3.0.1
## [13] ProjectSVR_0.2.0
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.3 spatstat.explore_3.2-3 reticulate_1.31
## [4] tidyselect_1.2.0 mlr3learners_0.5.6 htmlwidgets_1.6.2
## [7] BiocParallel_1.28.3 grid_4.1.2 Rtsne_0.16
## [10] mlr3misc_0.12.0 munsell_0.5.0 codetools_0.2-18
## [13] bbotk_0.7.2 ragg_1.2.5 ica_1.0-3
## [16] future_1.33.0 miniUI_0.1.1.1 mlr3verse_0.2.8
## [19] withr_2.5.0 spatstat.random_3.1-6 colorspace_2.1-0
## [22] progressr_0.14.0 highr_0.10 knitr_1.43
## [25] uuid_1.1-1 rstudioapi_0.15.0 stats4_4.1.2
## [28] ROCR_1.0-11 robustbase_0.99-0 tensor_1.5
## [31] listenv_0.9.0 labeling_0.4.3 mlr3tuning_0.19.0
## [34] polyclip_1.10-4 lgr_0.4.4 farver_2.1.1
## [37] rprojroot_2.0.3 parallelly_1.36.0 vctrs_0.6.3
## [40] generics_0.1.3 xfun_0.40 timechange_0.2.0
## [43] diptest_0.76-0 R6_2.5.1 doParallel_1.0.17
## [46] clue_0.3-64 flexmix_2.3-19 spatstat.utils_3.0-3
## [49] cachem_1.0.8 promises_1.2.1 scales_1.2.1
## [52] nnet_7.3-17 gtable_0.3.4 globals_0.16.2
## [55] goftest_1.2-3 mlr3hyperband_0.4.5 mlr3mbo_0.2.1
## [58] rlang_1.1.1 systemfonts_1.0.4 GlobalOptions_0.1.2
## [61] splines_4.1.2 lazyeval_0.2.2 paradox_0.11.1
## [64] spatstat.geom_3.2-5 checkmate_2.2.0 yaml_2.3.7
## [67] reshape2_1.4.4 abind_1.4-5 mlr3_0.16.1
## [70] backports_1.4.1 httpuv_1.6.11 tools_4.1.2
## [73] ellipsis_0.3.2 jquerylib_0.1.4 RColorBrewer_1.1-3
## [76] BiocGenerics_0.40.0 ggridges_0.5.4 Rcpp_1.0.11
## [79] plyr_1.8.8 deldir_1.0-9 pbapply_1.7-2
## [82] GetoptLong_1.0.5 cowplot_1.1.1 S4Vectors_0.32.4
## [85] zoo_1.8-12 ggrepel_0.9.3 cluster_2.1.2
## [88] here_1.0.1 fs_1.6.3 magrittr_2.0.3
## [91] data.table_1.14.8 scattermore_1.2 circlize_0.4.15
## [94] lmtest_0.9-40 RANN_2.6.1 fitdistrplus_1.1-11
## [97] matrixStats_1.0.0 stringfish_0.15.8 qs_0.25.5
## [100] hms_1.1.3 patchwork_1.1.3 mime_0.12
## [103] evaluate_0.21 xtable_1.8-4 mclust_6.0.0
## [106] IRanges_2.28.0 gridExtra_2.3 shape_1.4.6
## [109] UCell_1.3.1 compiler_4.1.2 mlr3cluster_0.1.8
## [112] KernSmooth_2.23-20 crayon_1.5.2 htmltools_0.5.6
## [115] tzdb_0.4.0 later_1.3.1 RcppParallel_5.1.7
## [118] RApiSerialize_0.1.2 ComplexHeatmap_2.10.0 rappdirs_0.3.3
## [121] MASS_7.3-55 fpc_2.2-10 mlr3data_0.7.0
## [124] Matrix_1.6-1 cli_3.6.1 parallel_4.1.2
## [127] igraph_1.5.1 pkgconfig_2.0.3 pkgdown_2.0.7
## [130] sp_2.0-0 plotly_4.10.2 spatstat.sparse_3.0-2
## [133] foreach_1.5.2 bslib_0.5.1 mlr3fselect_0.11.0
## [136] digest_0.6.33 sctransform_0.3.5 RcppAnnoy_0.0.21
## [139] mlr3filters_0.7.1 spatstat.data_3.0-1 rmarkdown_2.24
## [142] leiden_0.4.3 uwot_0.1.16 kernlab_0.9-32
## [145] shiny_1.7.5 modeltools_0.2-23 rjson_0.2.21
## [148] nlme_3.1-155 lifecycle_1.0.3 jsonlite_1.8.7
## [151] mlr3tuningspaces_0.4.0 desc_1.4.2 viridisLite_0.4.2
## [154] fansi_1.0.4 pillar_1.9.0 ggsci_3.0.0
## [157] lattice_0.20-45 fastmap_1.1.1 httr_1.4.7
## [160] DEoptimR_1.1-2 survival_3.2-13 glue_1.6.2
## [163] mlr3viz_0.6.1 png_0.1-8 prabclus_2.3-2
## [166] iterators_1.0.14 spacefillr_0.3.2 class_7.3-20
## [169] stringi_1.7.12 sass_0.4.7 mlr3pipelines_0.5.0-1
## [172] palmerpenguins_0.1.1 textshaping_0.3.6 memoise_2.0.1
## [175] irlba_2.3.5.1 future.apply_1.11.0