Build reference model for DISCO blood atlas
Compiled: November 06, 2023
Source:vignettes/model_pbmc.Rmd
model_pbmc.RmdIn this tutorial, we showed how to build the reference model for
blood atlas from DISCO
database. This case showed that once the reference atlas is well
annotated and integrated into a harmonious low dimensional embeddings,
we can easily training a supported vector regression model from gene set
scores to representated embeddings via ProjectSVR.
Download Reference Atlas
library(ProjectSVR)
library(Seurat)
library(tidyverse)
options(timeout = max(3600, getOption("timeout")))
`%notin%` <- Negate(`%in%`)
if (!dir.exists("reference")) dir.create("reference")
download.file(url = "https://zenodo.org/record/8350746/files/DISCO_hPBMCs.seurat.slim.qs",
destfile = "reference/DISCO_hPBMCs.seurat.slim.qs")Build Reference Model
data("pals")
seu.ref <- qs::qread("reference/DISCO_hPBMCs.seurat.slim.qs")
p1 <- DimPlot(seu.ref, pt.size = .4) + scale_color_manual(values = pals$disco_blood)
LabelClusters(p1, id = "ident")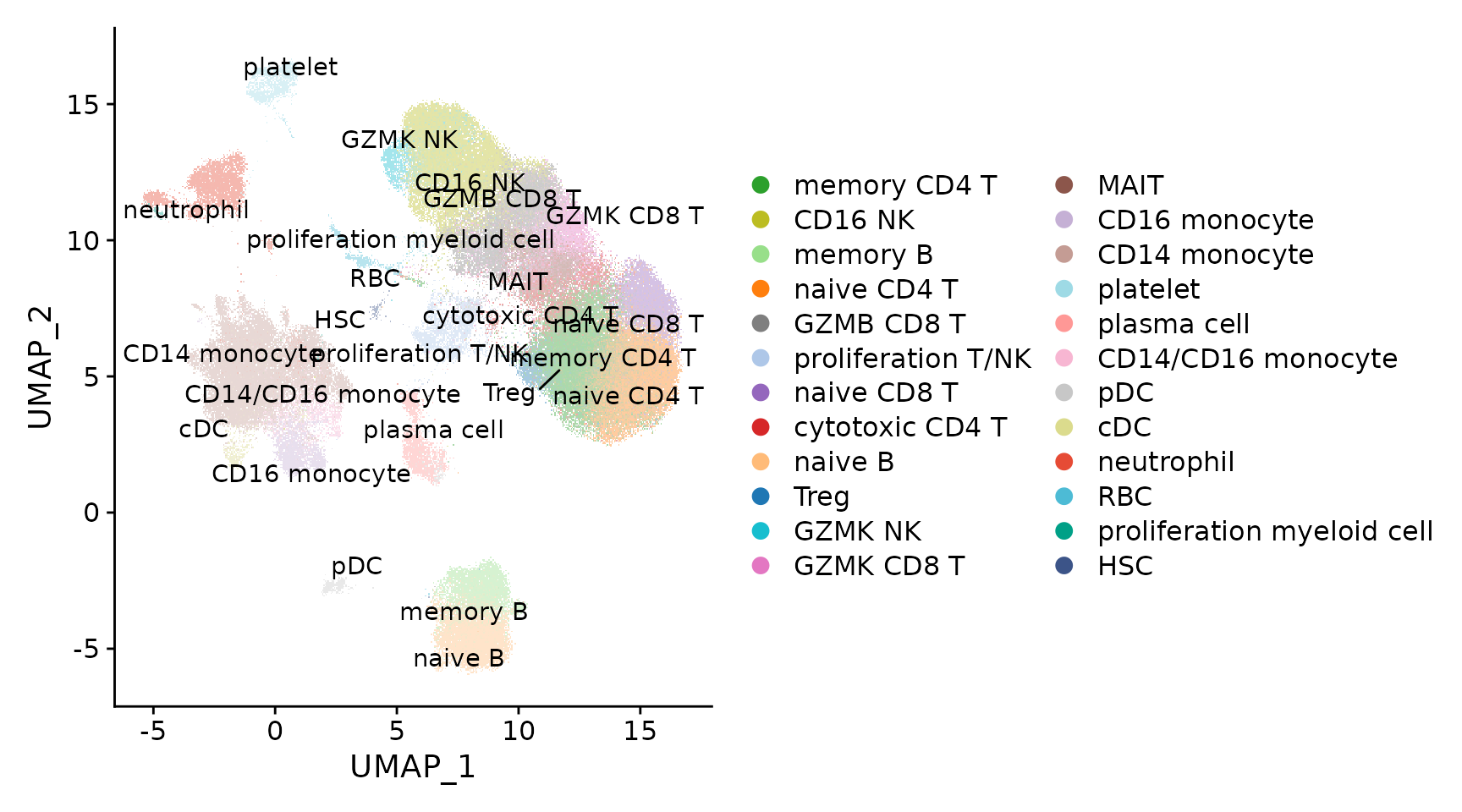
Extract signatures
Here we extract the top25 marker genes for each cell type (ribosomal and mitochondrial genes were removed).
##
## memory CD4 T CD16 NK
## 22151 22217
## memory B naive CD4 T
## 6413 20278
## GZMB CD8 T proliferation T/NK
## 13628 4058
## naive CD8 T cytotoxic CD4 T
## 7465 5956
## naive B Treg
## 9303 2881
## GZMK NK GZMK CD8 T
## 2298 7183
## MAIT CD16 monocyte
## 3542 2986
## CD14 monocyte platelet
## 23592 1649
## plasma cell CD14/CD16 monocyte
## 2601 1846
## pDC cDC
## 693 681
## neutrophil RBC
## 5245 710
## proliferation myeloid cell HSC
## 110 108
## To accelerate the calculation, we downsampled 200 cells from each cluster
seu.ref.ds <- subset(seu.ref, downsample = 200)
## The raw count matrix stores in `data` slot, so we copy it to `counts` slot for normalization.
seu.ref.ds[["RNA"]]@counts <- seu.ref.ds[["RNA"]]@data
seu.ref.ds <- NormalizeData(seu.ref.ds)
## Parallel calculation of the cell markers.
all.markers <- mcFindAllMarkers(seu.ref.ds, do.flatten = F, n.cores = 10)
top.genes <- lapply(all.markers, function(xx){
yy <- subset(xx, p_val_adj < 1e-6 & avg_log2FC > log2(1.5))
yy <- subset(yy, Gene.name.uniq %notin% ribo.genes)
yy <- yy[!grepl("^MT-", yy$Gene.name.uniq), ]
head(yy$Gene.name.uniq, 25)
})
sapply(top.genes, length)## memory CD4 T CD16 NK
## 25 25
## memory B naive CD4 T
## 25 25
## GZMB CD8 T proliferation T/NK
## 25 25
## naive CD8 T cytotoxic CD4 T
## 25 25
## naive B Treg
## 25 25
## GZMK NK GZMK CD8 T
## 25 25
## MAIT CD16 monocyte
## 25 25
## CD14 monocyte platelet
## 25 25
## plasma cell CD14/CD16 monocyte
## 25 25
## pDC cDC
## 25 25
## neutrophil RBC
## 25 25
## proliferation myeloid cell HSC
## 25 25Transfer raw count matrix to gene set score matrix
seu.ref <- ComputeModuleScore(seu.ref, gene.sets = top.genes, bg.genes = bg.genes,
method = "UCell", cores = 5)
# The signature score matrix is stored in 'SignatureScore' assay
Assays(seu.ref)## [1] "RNA" "SignatureScore"
DefaultAssay(seu.ref) <- "SignatureScore"Training reference model
gss.mat <- FetchData(seu.ref, vars = rownames(seu.ref))
embeddings.df <- FetchData(seu.ref, vars = paste0("UMAP_", 1:2))
batch.size = 8000 # number of subsampled cells for each SVR model
n.models = 20 # number of SVR models trained
umap.model <- FitEnsembleSVM(feature.mat = gss.mat,
emb.mat = embeddings.df,
batch.size = batch.size,
n.models = n.models,
cores = 10)Create reference object
meta.data: cell meta data (embeddings & cell type information)
gss.method: method used in
ComputeModuleScore()[optional] colors: for plots
[optional] text.pos: text annotation on the reference plots
meta.data <- FetchData(seu.ref, vars = c(paste0("UMAP_", 1:2), "cell_type", "cell_subtype") )
colors <- pals$disco_blood
text.pos <- data.frame(
x = c(11, 2, 6, -2, 0, 13, 10, 14, 7, -2, 1, 3, 4, 7),
y = c(-4, -4, 1, 1, 5, 2, 4, 12, 16, 12, 15, 9, 6.5, 5),
label = c("B", "pDC", "plasma", "cDC", "mono", "CD4 T", "Treg", "CD8 T",
"NK", "neutrophil", "platelet", "RBC", "HSC", "T/NK pro")
)
reference <- CreateReference(umap.model = umap.model,
gene.sets = top.genes,
bg.genes = bg.genes,
meta.data = meta.data,
gss.method = "UCell",
colors = colors,
text.pos = text.pos)
dir.create("models")
saveRDS(reference, "models/model.disco_pbmc.rds")Session Info
## R version 4.1.2 (2021-11-01)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3
## [5] purrr_1.0.2 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
## [9] ggplot2_3.4.3 tidyverse_2.0.0 SeuratObject_4.1.3 Seurat_4.3.0.1
## [13] ProjectSVR_0.2.0
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.3 spatstat.explore_3.2-3 reticulate_1.31
## [4] tidyselect_1.2.0 mlr3learners_0.5.6 htmlwidgets_1.6.2
## [7] BiocParallel_1.28.3 grid_4.1.2 Rtsne_0.16
## [10] mlr3misc_0.12.0 munsell_0.5.0 codetools_0.2-18
## [13] bbotk_0.7.2 ragg_1.2.5 ica_1.0-3
## [16] future_1.33.0 miniUI_0.1.1.1 mlr3verse_0.2.8
## [19] withr_2.5.0 spatstat.random_3.1-6 colorspace_2.1-0
## [22] progressr_0.14.0 highr_0.10 knitr_1.43
## [25] uuid_1.1-1 rstudioapi_0.15.0 stats4_4.1.2
## [28] ROCR_1.0-11 robustbase_0.99-0 tensor_1.5
## [31] listenv_0.9.0 labeling_0.4.3 mlr3tuning_0.19.0
## [34] polyclip_1.10-4 lgr_0.4.4 farver_2.1.1
## [37] rprojroot_2.0.3 parallelly_1.36.0 vctrs_0.6.3
## [40] generics_0.1.3 xfun_0.40 timechange_0.2.0
## [43] diptest_0.76-0 R6_2.5.1 doParallel_1.0.17
## [46] clue_0.3-64 flexmix_2.3-19 spatstat.utils_3.0-3
## [49] cachem_1.0.8 promises_1.2.1 scales_1.2.1
## [52] nnet_7.3-17 gtable_0.3.4 globals_0.16.2
## [55] goftest_1.2-3 mlr3hyperband_0.4.5 mlr3mbo_0.2.1
## [58] rlang_1.1.1 systemfonts_1.0.4 GlobalOptions_0.1.2
## [61] splines_4.1.2 lazyeval_0.2.2 paradox_0.11.1
## [64] spatstat.geom_3.2-5 checkmate_2.2.0 yaml_2.3.7
## [67] reshape2_1.4.4 abind_1.4-5 mlr3_0.16.1
## [70] backports_1.4.1 httpuv_1.6.11 tools_4.1.2
## [73] ellipsis_0.3.2 jquerylib_0.1.4 RColorBrewer_1.1-3
## [76] BiocGenerics_0.40.0 ggridges_0.5.4 Rcpp_1.0.11
## [79] plyr_1.8.8 deldir_1.0-9 pbapply_1.7-2
## [82] GetoptLong_1.0.5 cowplot_1.1.1 S4Vectors_0.32.4
## [85] zoo_1.8-12 ggrepel_0.9.3 cluster_2.1.2
## [88] fs_1.6.3 magrittr_2.0.3 data.table_1.14.8
## [91] scattermore_1.2 circlize_0.4.15 lmtest_0.9-40
## [94] RANN_2.6.1 fitdistrplus_1.1-11 matrixStats_1.0.0
## [97] stringfish_0.15.8 qs_0.25.5 hms_1.1.3
## [100] patchwork_1.1.3 mime_0.12 evaluate_0.21
## [103] xtable_1.8-4 mclust_6.0.0 IRanges_2.28.0
## [106] gridExtra_2.3 shape_1.4.6 UCell_1.3.1
## [109] compiler_4.1.2 mlr3cluster_0.1.8 KernSmooth_2.23-20
## [112] crayon_1.5.2 htmltools_0.5.6 tzdb_0.4.0
## [115] later_1.3.1 RcppParallel_5.1.7 RApiSerialize_0.1.2
## [118] ComplexHeatmap_2.10.0 MASS_7.3-55 fpc_2.2-10
## [121] mlr3data_0.7.0 Matrix_1.6-1 cli_3.6.1
## [124] parallel_4.1.2 igraph_1.5.1 pkgconfig_2.0.3
## [127] pkgdown_2.0.7 sp_2.0-0 plotly_4.10.2
## [130] spatstat.sparse_3.0-2 foreach_1.5.2 bslib_0.5.1
## [133] mlr3fselect_0.11.0 digest_0.6.33 sctransform_0.3.5
## [136] RcppAnnoy_0.0.21 mlr3filters_0.7.1 spatstat.data_3.0-1
## [139] rmarkdown_2.24 leiden_0.4.3 uwot_0.1.16
## [142] kernlab_0.9-32 shiny_1.7.5 modeltools_0.2-23
## [145] rjson_0.2.21 nlme_3.1-155 lifecycle_1.0.3
## [148] jsonlite_1.8.7 mlr3tuningspaces_0.4.0 desc_1.4.2
## [151] viridisLite_0.4.2 fansi_1.0.4 pillar_1.9.0
## [154] lattice_0.20-45 fastmap_1.1.1 httr_1.4.7
## [157] DEoptimR_1.1-2 survival_3.2-13 glue_1.6.2
## [160] mlr3viz_0.6.1 png_0.1-8 prabclus_2.3-2
## [163] iterators_1.0.14 spacefillr_0.3.2 class_7.3-20
## [166] stringi_1.7.12 sass_0.4.7 mlr3pipelines_0.5.0-1
## [169] palmerpenguins_0.1.1 textshaping_0.3.6 memoise_2.0.1
## [172] irlba_2.3.5.1 future.apply_1.11.0