Accurate identification of the tumor-infiltrated T cell heterogeneity in cancer immunology study
Compiled: November 06, 2023
Source:vignettes/mapQuery_ICB_BRCA.Rmd
mapQuery_ICB_BRCA.RmdIn recent years, single-cell RNA-sequencing (scRNA-seq) enabled unbiased exploration of T cell diversity in health, disease and response to therapies at an unprecedented scale and resolution. However, a comprehensive definition of T cell “reference” subtypes remains elusive 1.
Several groups try to integrate T cells in the tumor micro-environment (TME) across different cancer types to build a pan-cancer T cell landscape. For example, Zheng et al. depicted a pan-cancer landscape of T cell heterogeneity in the TME and established a baseline reference for future scRNA-seq studies associated with cancer treatments 2.
In this tutorial, we demonstrate the process of projecting query T cells onto the pan-cancer T cell landscape (PTCL) to interpret the disparities in CD8 T cell heterogeneity between responsive and non-responsive patients with breast cancer who have undergone immune checkpoint blockade (ICB) therapy 3. This will be done using ProjectSVR.
Download Related Dataset
library(Seurat)
library(ProjectSVR)
library(tidyverse)
options(timeout = max(3600, getOption("timeout")))
# reference model
if (!dir.exists("models")) dir.create("models")
download.file(url = "https://zenodo.org/record/8350732/files/model.Zheng2021.CD8Tcell.rds",
destfile = "models/model.Zheng2021.CD8Tcell.rds")
# query data
if (!dir.exists("query")) dir.create("query")
download.file(url = "https://zenodo.org/record/8350748/files/ICB_BRCA.1864-counts_cd8tcell_cohort1.seurat.rds",
destfile = "query/ICB_BRCA.1864-counts_cd8tcell_cohort1.seurat.rds")Map Query to Reference
Reference mapping
reference <- readRDS("models/model.Zheng2021.CD8Tcell.rds")
seu.q <- readRDS("query/ICB_BRCA.1864-counts_cd8tcell_cohort1.seurat.rds")
annotations <- c(
"E" = "Responders",
"n/a" = "n/a",
"NE" = "Non-Responders"
)
seu.q$group <- annotations[seu.q$expansion]
seu.q <- ProjectSVR::MapQuery(seu.q, reference = reference,
add.map.qual = T, ncores = 10)
p1 <- DimPlot(seu.q, reduction = "ref.umap", group.by = "patient_id")
p2 <- DimPlot(seu.q, reduction = "ref.umap", group.by = "cellSubType",
label = T) + ggsci::scale_color_d3("category20")
p1 + p2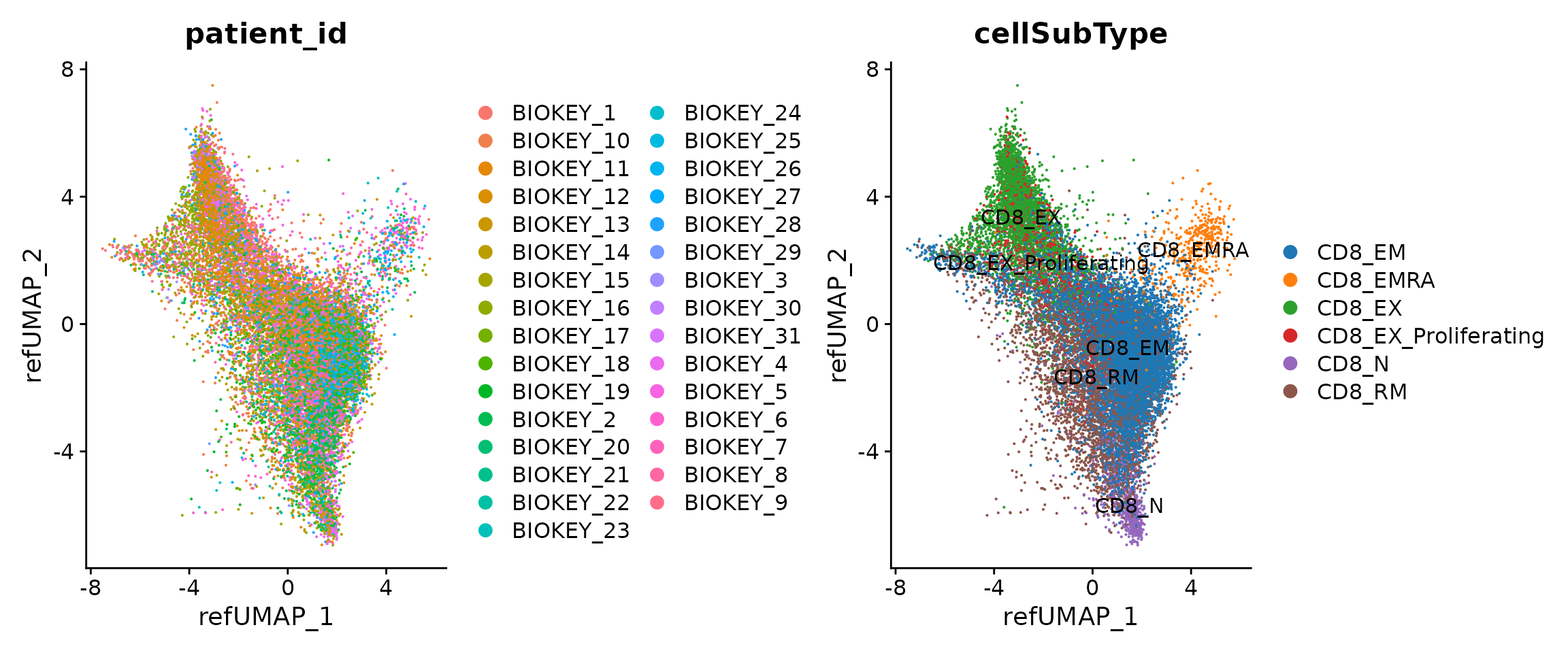
Maping quality
Determine the threshold to distinguish the good and bad projections.
QC plot for the reference mapping.
## cutoff by adjusted p value
MapQCPlot(seu.q, p.adj.cutoff = 1e-3)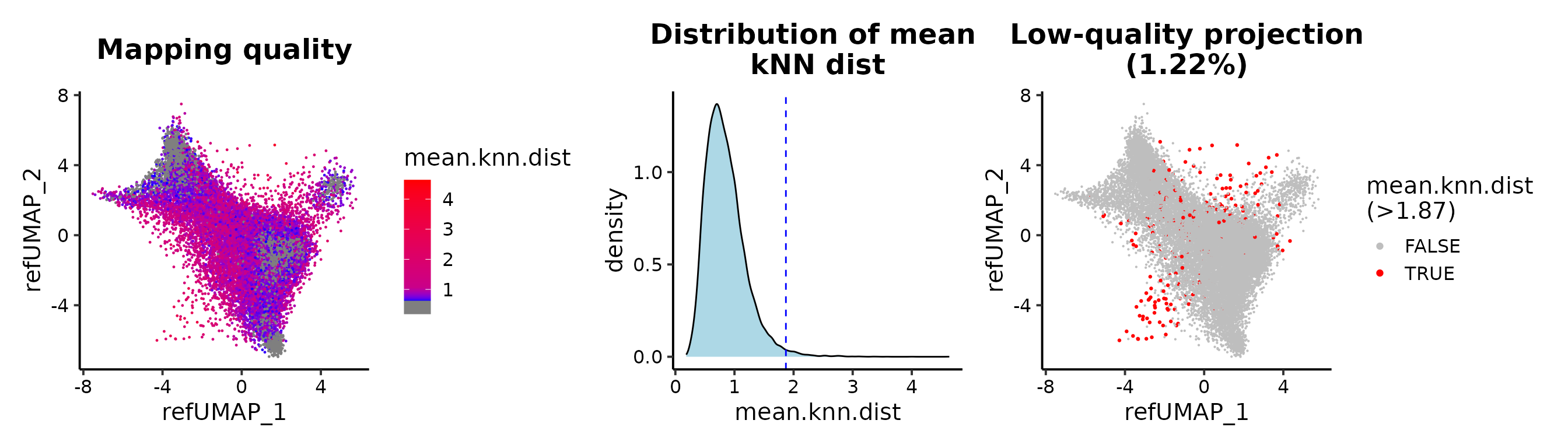
## or mean.knn.dist
MapQCPlot(seu.q, map.q.cutoff = 1.6)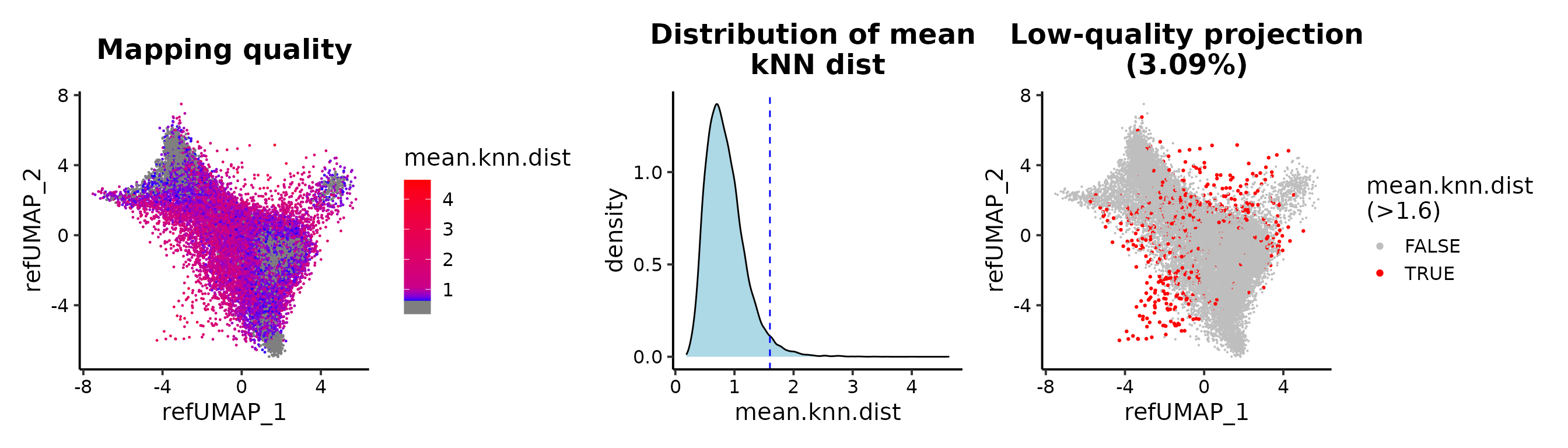
Visualize the projected query cells onto the reference atlas.
PlotProjection(seu.q, reference, split.by = "cellSubType", ref.color.by = "cluster.name",
ref.size = .5, ref.alpha = .3, query.size = 1, query.alpha = .5, n.row = 2)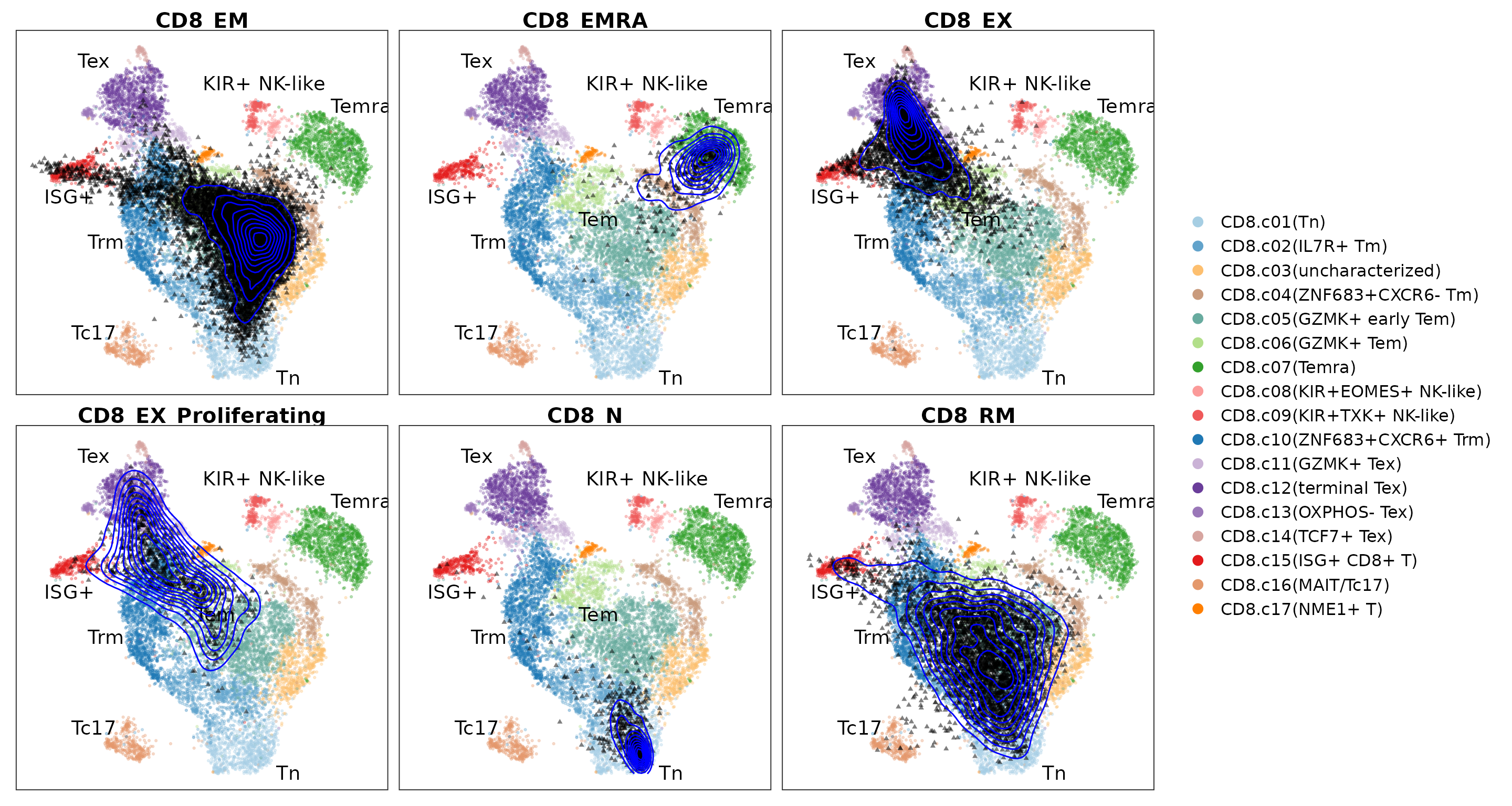
Label transfer
seu.q <- ProjectSVR::LabelTransfer(seu.q, reference, ref.label.col = "cluster.name")Differential cell population
seu.q <- subset(seu.q, group != "n/a" & timepoint == "Pre")
seu.q <- subset(seu.q, mean.knn.dist < 1.6)
seu.q$group <- factor(seu.q$group)
PlotProjection(seu.q, reference, split.by = "group", ref.color.by = "cluster.name",
ref.size = .5, ref.alpha = .3, query.size = 1, query.alpha = .5, n.row = 1)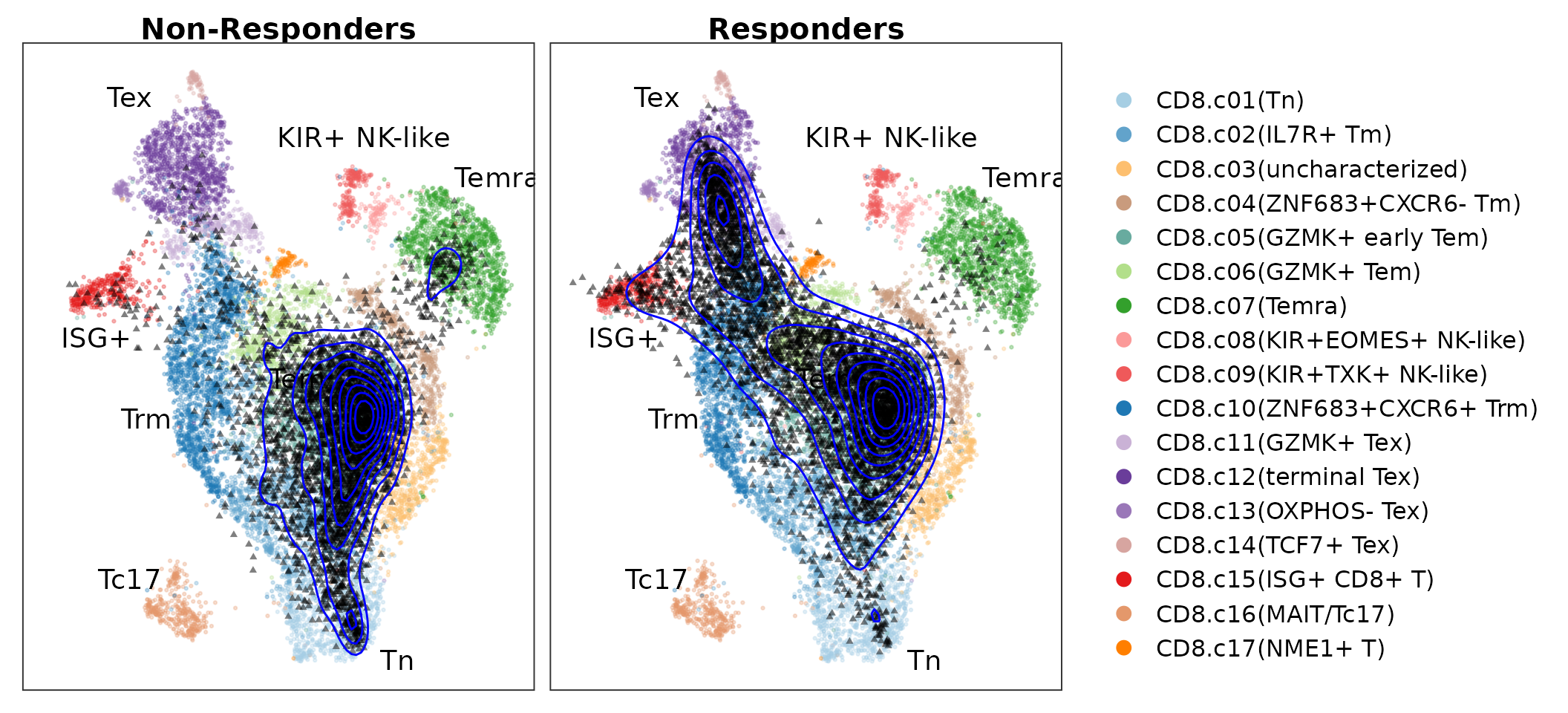
Alluvia plot
AlluviaPlot(seu.q@meta.data, by = "group", fill = "knn.pred.celltype",
colors = reference$ref.cellmeta$colors,
bar.width = .5)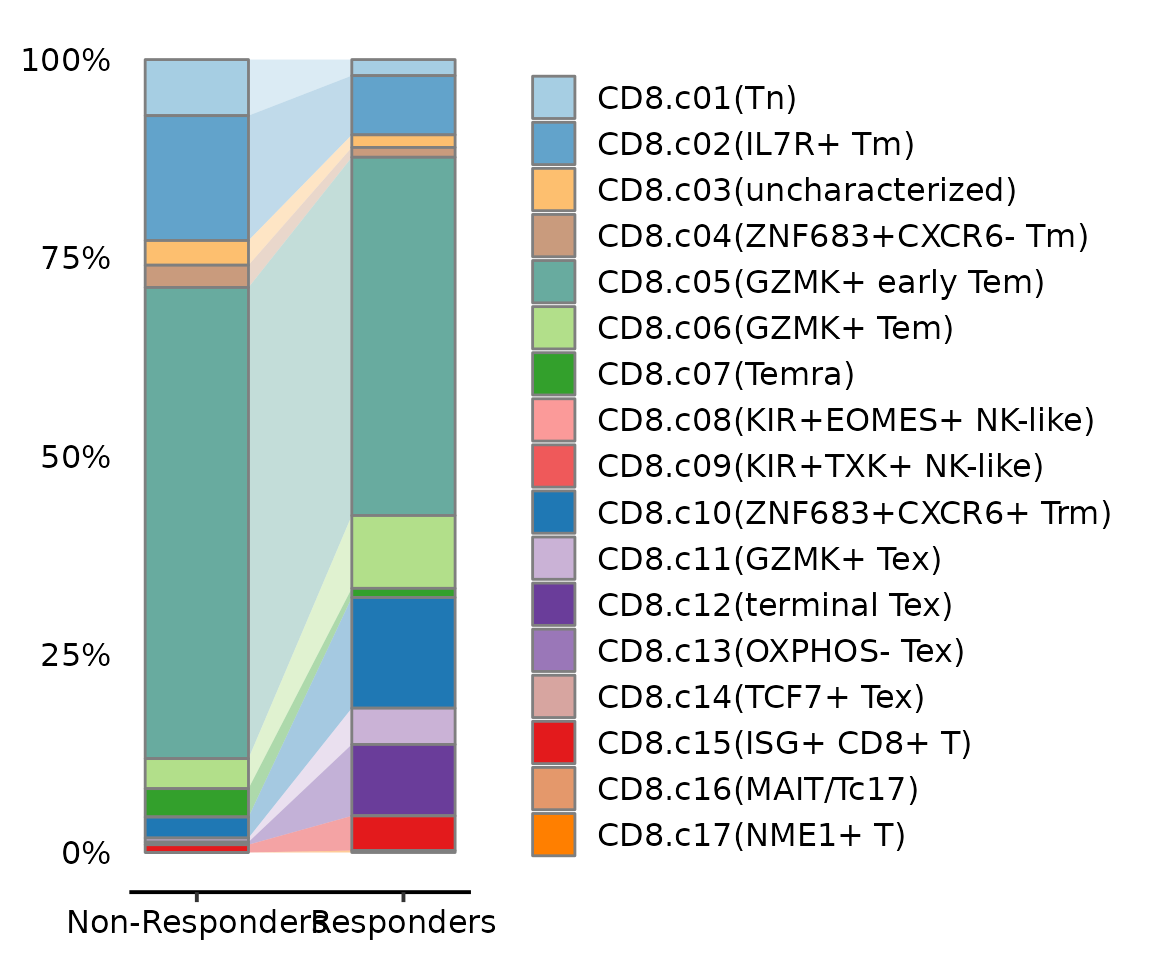
Preference analysis
GroupPreferencePlot(seu.q@meta.data, group.by = "group",
preference.on = "knn.pred.celltype",
column_names_rot = 0, column_names_centered = TRUE)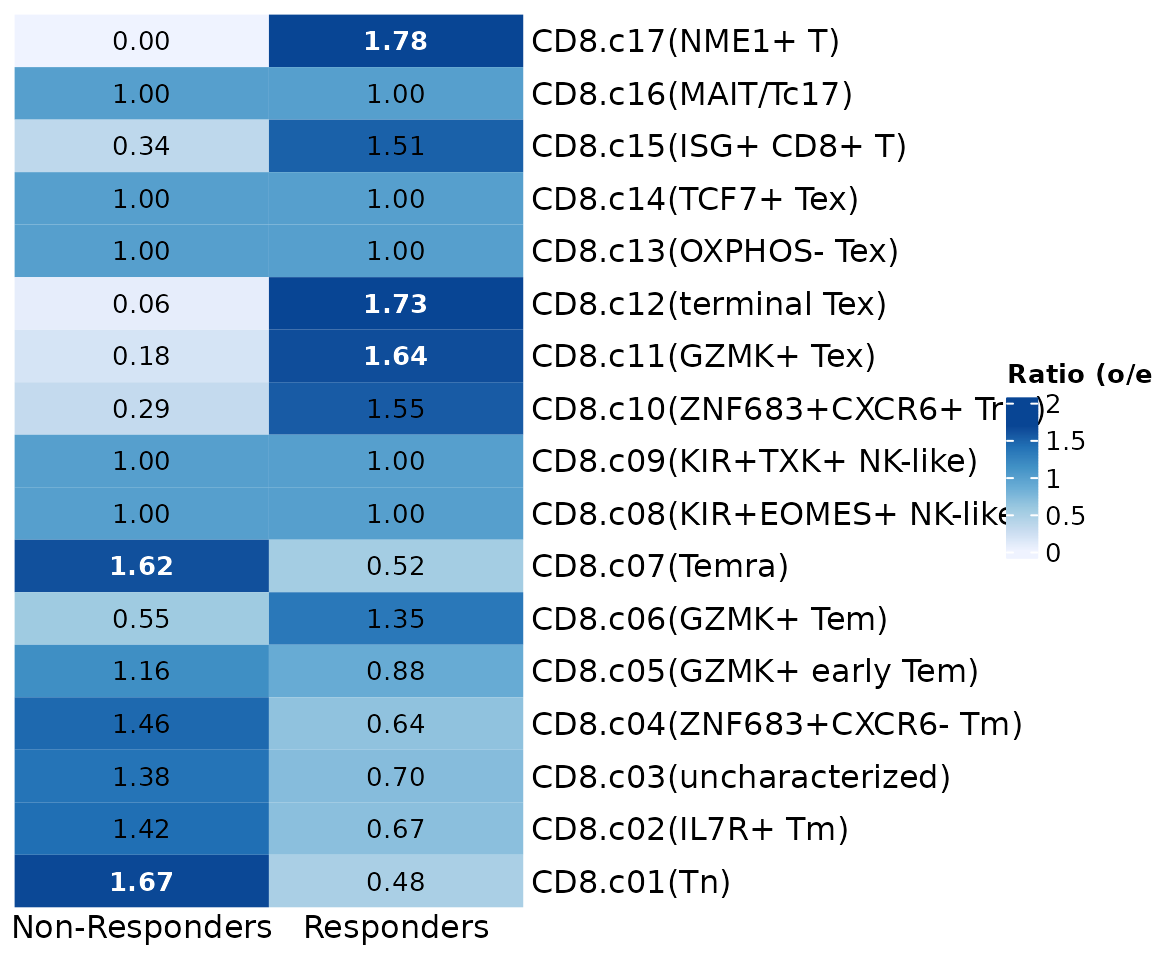
If seu.q$knn.pred.celltype was not a factor, then rows
were ordered by o/e value.
meta.data <- seu.q@meta.data
meta.data$knn.pred.celltype <- as.character(meta.data$knn.pred.celltype)
GroupPreferencePlot(meta.data, group.by = "group",
preference.on = "knn.pred.celltype",
column_names_rot = 0, column_names_centered = TRUE)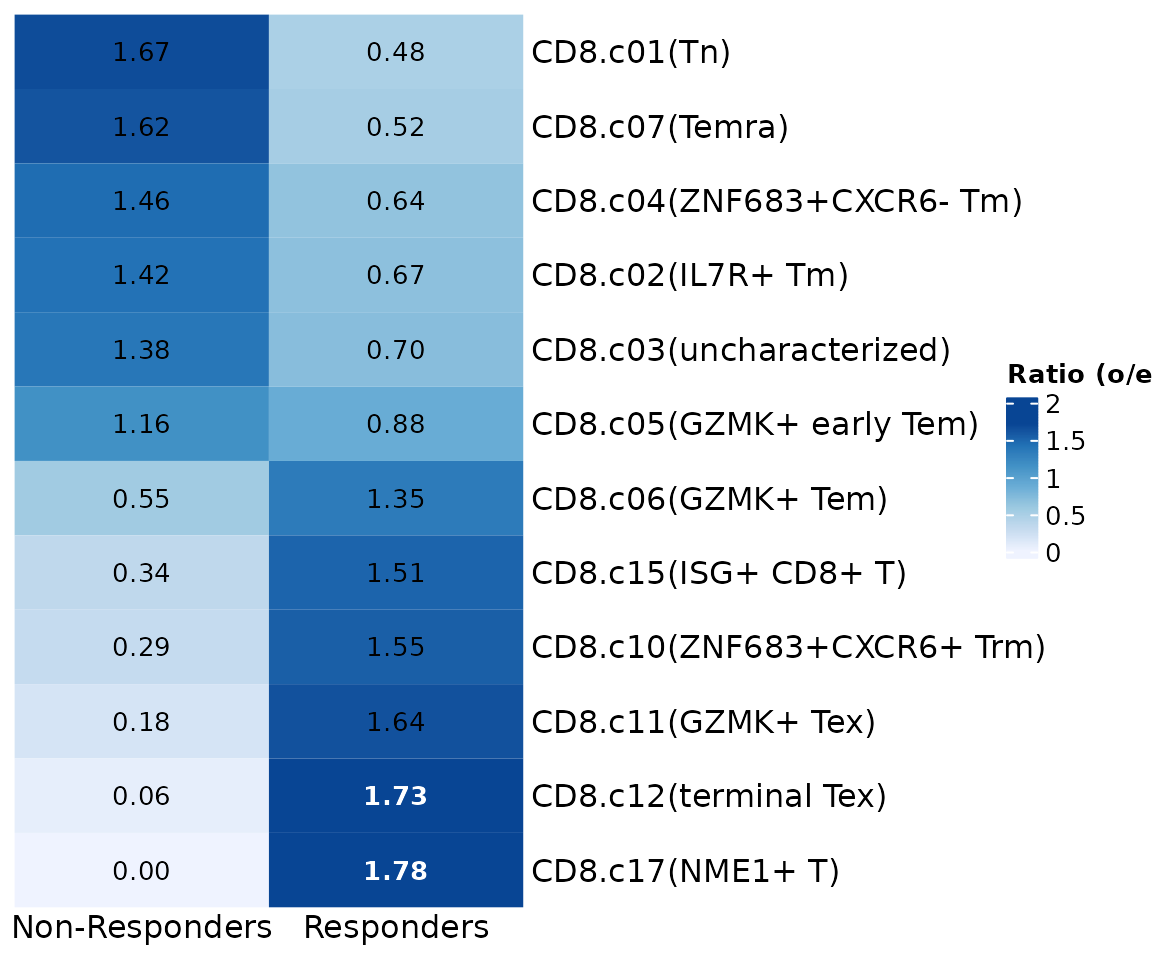
Wilcoxon test
da.test <- AbundanceTest(cellmeta = seu.q@meta.data,
celltype.col = "knn.pred.celltype",
sample.col = "patient_id",
group.col = "group")
## Volcano plot
VolcanoPlot(da.test, xlab = "log2(R / NR)", ylab = "Frequency",
colors = reference$ref.cellmeta$colors)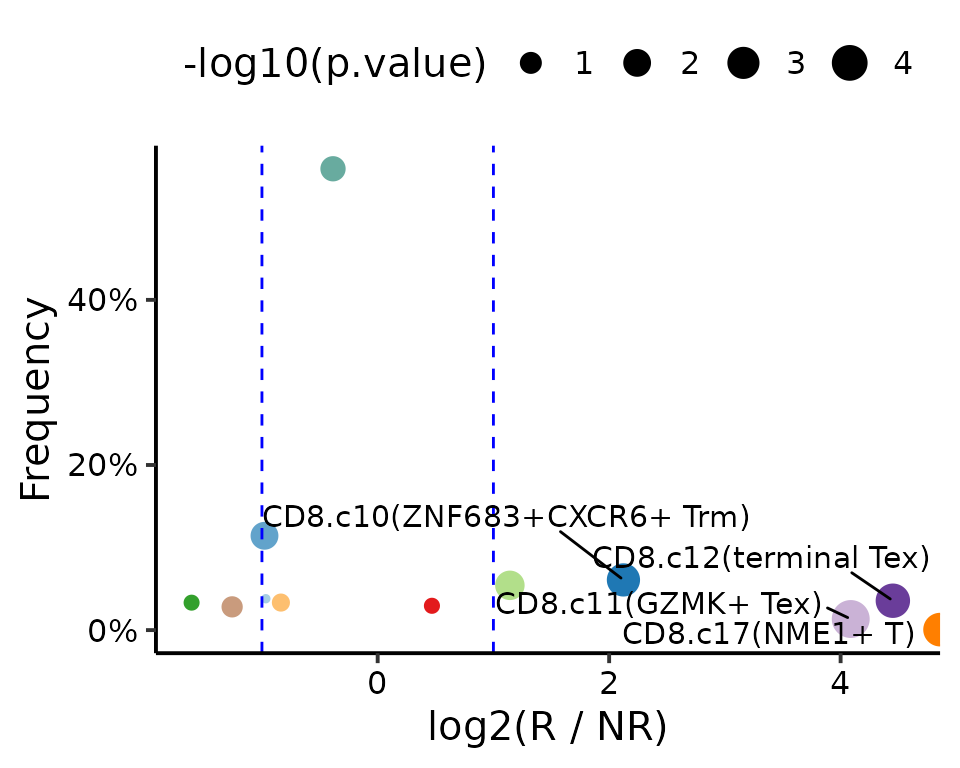
Focus on the detailed information via BoxPlot().
## Box plot
BoxPlot(cellmeta = seu.q@meta.data, celltype.col = "knn.pred.celltype",
sample.col = "patient_id", group.col = "group",
celltypes.show = c("CD8.c12(terminal Tex)", "CD8.c11(GZMK+ Tex)"),
legend.ncol = 2) +
scale_x_discrete(labels = c("NR", "R"))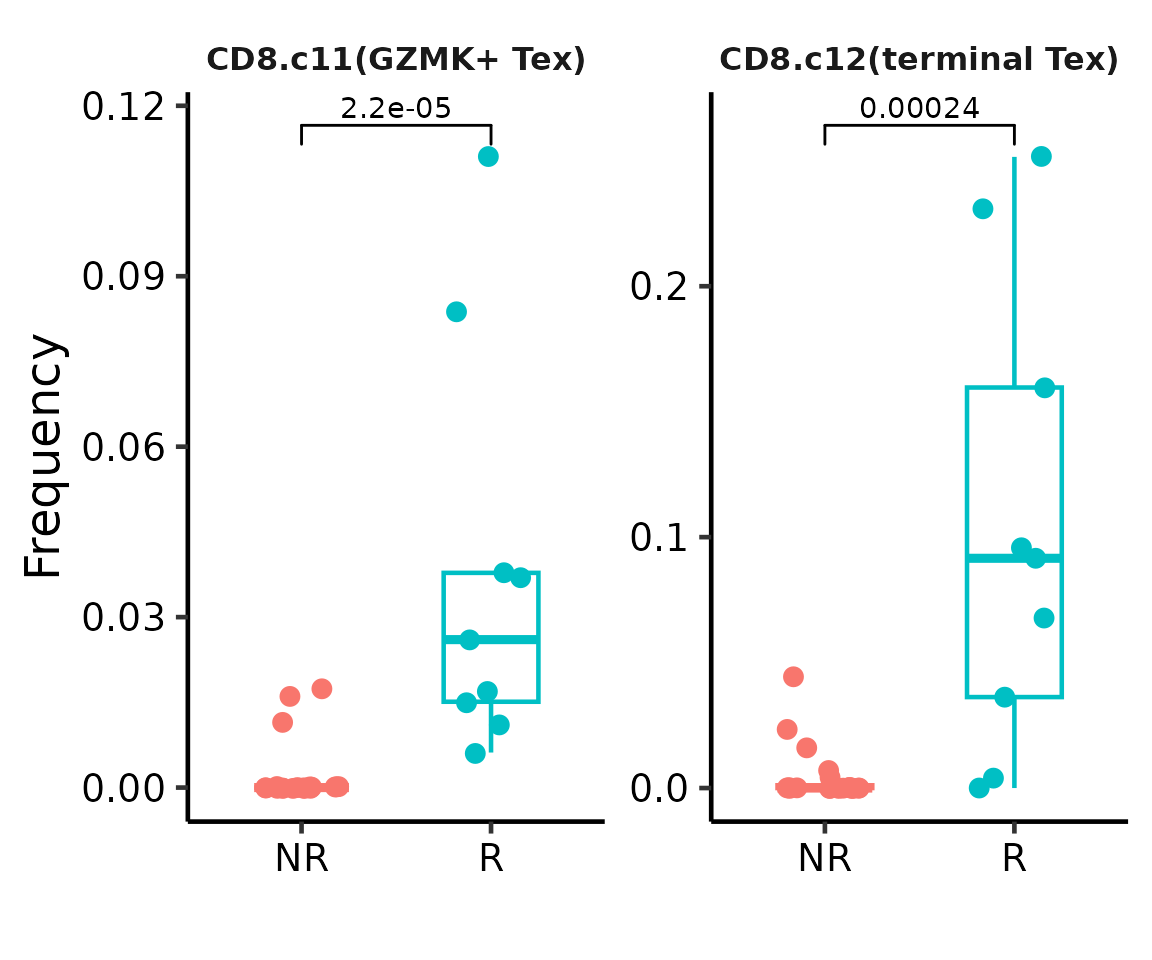
Session Info
## R version 4.1.2 (2021-11-01)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] lubridate_1.9.2 forcats_1.0.0 stringr_1.5.0 dplyr_1.1.3
## [5] purrr_1.0.2 readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
## [9] ggplot2_3.4.3 tidyverse_2.0.0 ProjectSVR_0.2.0 SeuratObject_4.1.3
## [13] Seurat_4.3.0.1
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.3 spatstat.explore_3.2-3 reticulate_1.31
## [4] tidyselect_1.2.0 mlr3learners_0.5.6 htmlwidgets_1.6.2
## [7] BiocParallel_1.28.3 grid_4.1.2 Rtsne_0.16
## [10] mlr3misc_0.12.0 munsell_0.5.0 codetools_0.2-18
## [13] bbotk_0.7.2 ragg_1.2.5 ica_1.0-3
## [16] future_1.33.0 miniUI_0.1.1.1 mlr3verse_0.2.8
## [19] withr_2.5.0 spatstat.random_3.1-6 colorspace_2.1-0
## [22] progressr_0.14.0 highr_0.10 knitr_1.43
## [25] uuid_1.1-1 rstudioapi_0.15.0 stats4_4.1.2
## [28] ROCR_1.0-11 robustbase_0.99-0 ggsignif_0.6.4
## [31] tensor_1.5 listenv_0.9.0 labeling_0.4.3
## [34] mlr3tuning_0.19.0 lgr_0.4.4 polyclip_1.10-4
## [37] farver_2.1.1 rprojroot_2.0.3 parallelly_1.36.0
## [40] vctrs_0.6.3 generics_0.1.3 xfun_0.40
## [43] timechange_0.2.0 diptest_0.76-0 R6_2.5.1
## [46] doParallel_1.0.17 clue_0.3-64 isoband_0.2.7
## [49] flexmix_2.3-19 spatstat.utils_3.0-3 cachem_1.0.8
## [52] promises_1.2.1 scales_1.2.1 nnet_7.3-17
## [55] gtable_0.3.4 globals_0.16.2 mlr3hyperband_0.4.5
## [58] goftest_1.2-3 mlr3mbo_0.2.1 rlang_1.1.1
## [61] systemfonts_1.0.4 GlobalOptions_0.1.2 splines_4.1.2
## [64] lazyeval_0.2.2 paradox_0.11.1 spatstat.geom_3.2-5
## [67] checkmate_2.2.0 yaml_2.3.7 reshape2_1.4.4
## [70] abind_1.4-5 mlr3_0.16.1 backports_1.4.1
## [73] httpuv_1.6.11 tools_4.1.2 ellipsis_0.3.2
## [76] jquerylib_0.1.4 RColorBrewer_1.1-3 BiocGenerics_0.40.0
## [79] ggridges_0.5.4 Rcpp_1.0.11 plyr_1.8.8
## [82] deldir_1.0-9 pbapply_1.7-2 GetoptLong_1.0.5
## [85] cowplot_1.1.1 S4Vectors_0.32.4 zoo_1.8-12
## [88] ggrepel_0.9.3 cluster_2.1.2 here_1.0.1
## [91] fs_1.6.3 magrittr_2.0.3 data.table_1.14.8
## [94] scattermore_1.2 circlize_0.4.15 lmtest_0.9-40
## [97] RANN_2.6.1 fitdistrplus_1.1-11 matrixStats_1.0.0
## [100] hms_1.1.3 patchwork_1.1.3 mime_0.12
## [103] evaluate_0.21 xtable_1.8-4 mclust_6.0.0
## [106] IRanges_2.28.0 gridExtra_2.3 shape_1.4.6
## [109] UCell_1.3.1 compiler_4.1.2 mlr3cluster_0.1.8
## [112] KernSmooth_2.23-20 crayon_1.5.2 htmltools_0.5.6
## [115] tzdb_0.4.0 later_1.3.1 ComplexHeatmap_2.10.0
## [118] rappdirs_0.3.3 MASS_7.3-55 fpc_2.2-10
## [121] mlr3data_0.7.0 Matrix_1.6-1 cli_3.6.1
## [124] parallel_4.1.2 igraph_1.5.1 pkgconfig_2.0.3
## [127] pkgdown_2.0.7 sp_2.0-0 plotly_4.10.2
## [130] spatstat.sparse_3.0-2 foreach_1.5.2 bslib_0.5.1
## [133] mlr3fselect_0.11.0 digest_0.6.33 sctransform_0.3.5
## [136] RcppAnnoy_0.0.21 mlr3filters_0.7.1 spatstat.data_3.0-1
## [139] rmarkdown_2.24 leiden_0.4.3 uwot_0.1.16
## [142] kernlab_0.9-32 shiny_1.7.5 modeltools_0.2-23
## [145] rjson_0.2.21 lifecycle_1.0.3 nlme_3.1-155
## [148] jsonlite_1.8.7 mlr3tuningspaces_0.4.0 desc_1.4.2
## [151] viridisLite_0.4.2 fansi_1.0.4 pillar_1.9.0
## [154] ggsci_3.0.0 lattice_0.20-45 fastmap_1.1.1
## [157] httr_1.4.7 DEoptimR_1.1-2 survival_3.2-13
## [160] glue_1.6.2 mlr3viz_0.6.1 png_0.1-8
## [163] prabclus_2.3-2 iterators_1.0.14 spacefillr_0.3.2
## [166] class_7.3-20 stringi_1.7.12 sass_0.4.7
## [169] mlr3pipelines_0.5.0-1 palmerpenguins_0.1.1 textshaping_0.3.6
## [172] memoise_2.0.1 irlba_2.3.5.1 future.apply_1.11.0